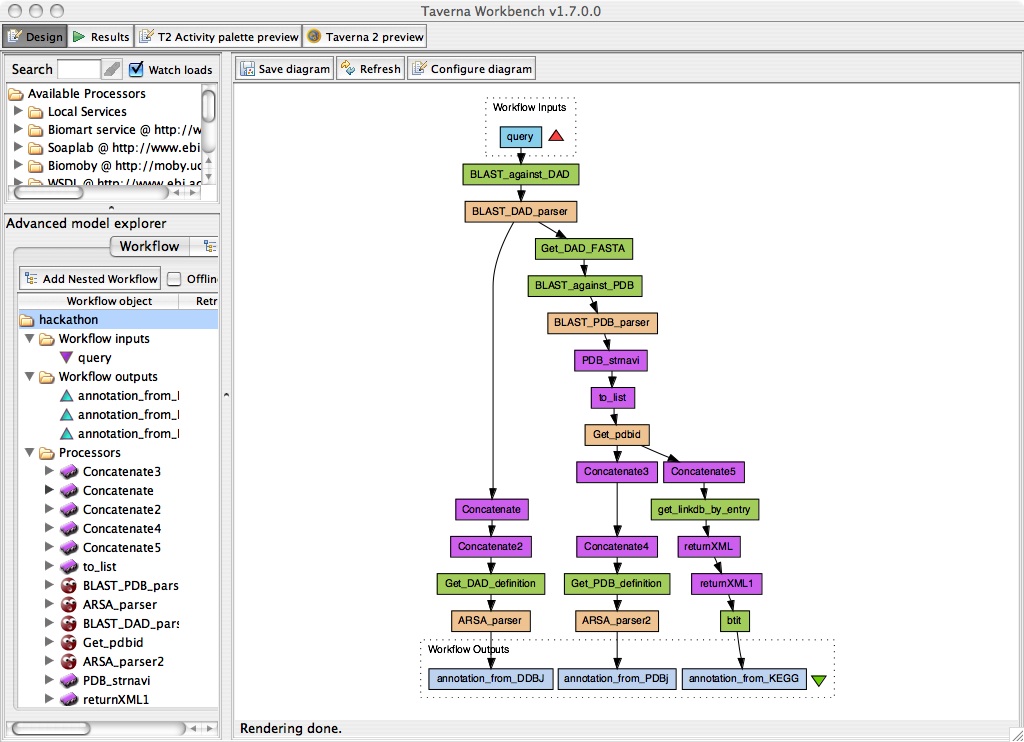
Summary of PDBj-DDBJ-KEGG workflow demo
get Taverna workflow from [PDBj-DDBJ-KEGG].
Who we are
Our Task
Somehow integrate DDBJ, PDBj, and KEGG.
After a lot of proposals were rejected, we decided to make a certain workflow to explore the possibilities and limitations and problems for (our) DB integration.
- By the way, the biggest problem was that we did not know any specific needs.
What it is
Annotate a protein sequence by homology based on sequence and structure.
Input: Given an unannotated protein sequence ("hypothetical", "putative", etc.)
- Run BLAST against DAD at DDBJ.
- When annotated homologs are found, get their annotations from DDBJ.
- When only hypothetical proteins are found, run BLAST against PDB.
- If any homologs are found (annotated or not), they are sent to Structural-Navigator (structure search) at PDBj.
- If any similar structures are found, get the annotations from PDBj and KEGG.
Output: homology-based annotation from DDBJ, KEGG, and PDBj
How we implemented it
- We used Taverna. (Thanks you, Stuart!)
- DDBJ and KEGG have plenty of SOAP services which are actually used.
- PDBj's SOAP services are very poor (no SOAP service for Structure-Navi), but there are some REST services which were used after some modifications.
- Only one of us knew Java (Mr. Shigemoto from DDBJ) so he did most of the programming (BeanShell scripts).
What we learned
- No conditional branch in Taverna.
- We needed to write many small glue codes to connect different services.
- BeanShell scripts are handy.
- But, writing BeanShell scripts requires a Java programmer (We had only Shigemoto-san...).
- SOAP is usable (AK's comment).
- It's nice to have knowledgeable people around. We can ask questions and modify codes instantly.
What can be improved
- Server-side programming?
- Lots of small widgets?
- Knowing the target users. (Who are they, anyway?)
Sort of conclusions…
- It's unrealistic to make strictly standardized data format and/or API.
- First of all, individual providers should hack to make usable services.
- Only after then, we can make interfaces interoperable by hacking.
- Thus, it's nice to have a Hackathon once in a while...
Attachments
-
DDBJ-KEGG-PDBj.jpg
 (190.8 KB) - added by akinjo
17 years ago.
(190.8 KB) - added by akinjo
17 years ago.